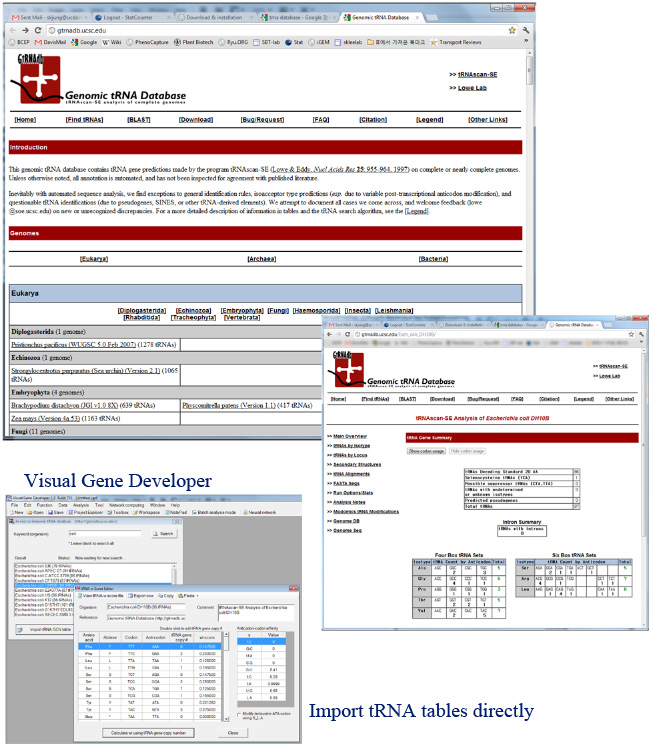
Web DB Access
In order to optimize codon or calculate CAI value, a user has to build a codon usage table. Visual Gene Developer can retrieve codon usage table data from Kazusa codon usage web database (http://www.kazusa.or.jp/codon/). First of all, we will explain 'codon usage table and w-value 'and then describe how to import codon usage table data from the web database.
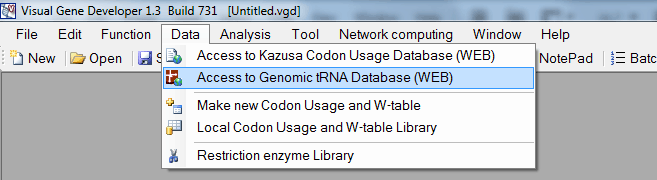
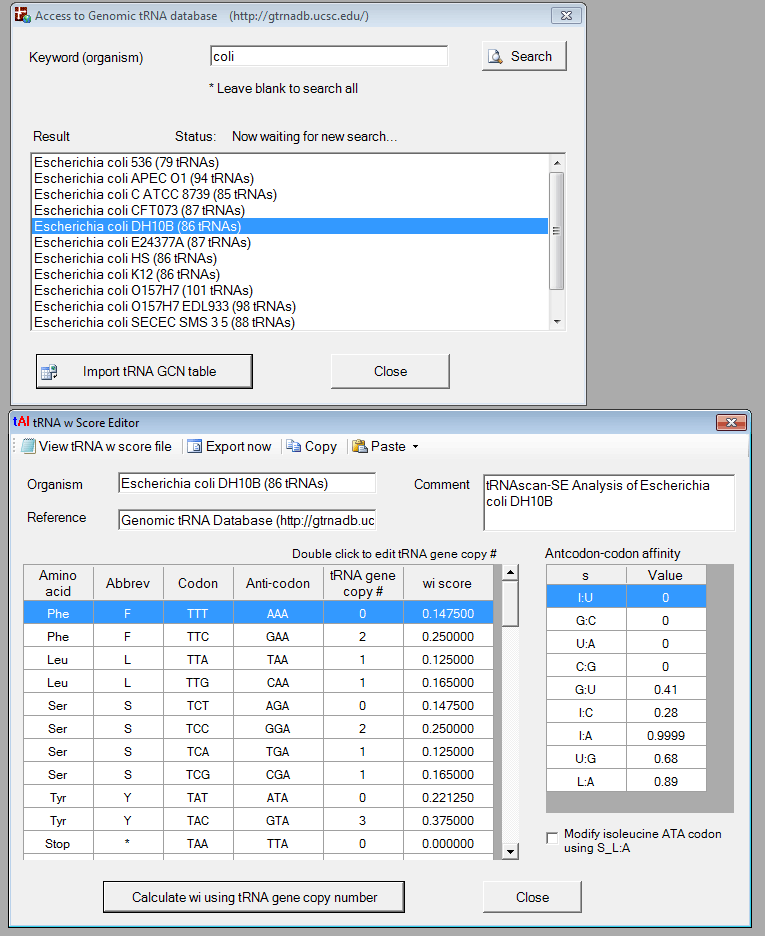
We also implemented a function that allows a user to access the Genomic tRNA database (http://gtrnadb.ucsc.edu/) and calculate tAI (tRNA adaptation index).
o Codon usage and w table
1. Click on the 'Codon Usage and W-table Library' in the 'Data' menu
2. You can see the new window, 'Codon usage and W-table Library'. Choose one of the files (for example, Citrus sinensis.vgw).
3. Click on the 'Edit' button
4. Again, a new window, 'Codon usage and w-table [file name]' will be shown. Please check the table below.
We simply call it 'Codon usage and w-table'.
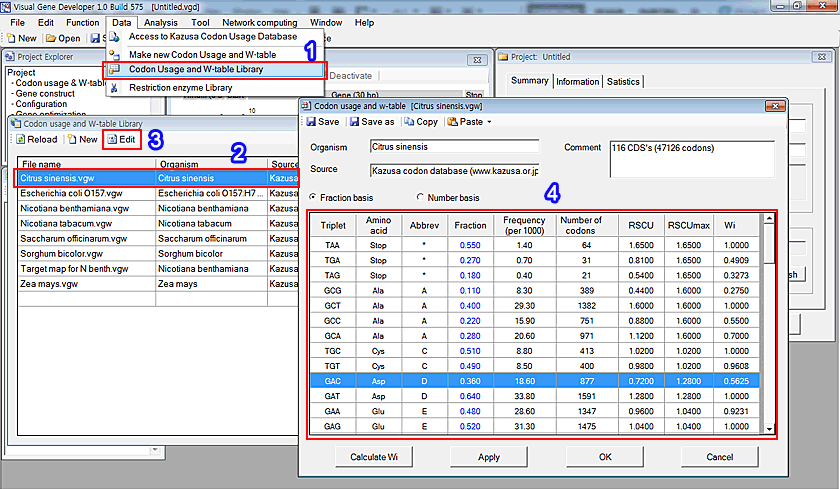
Among the parameters such as Fraction, Frequency, and RSCU, Wi is the most important parameter that is used to calculate CAI (Codon Adaptation Index). One of the modules, specifically 'Monte-Carlo Codon Optimization', also use it to optimize gene. Therefore, if you decide a target host organism or know the optimum codon usage table, first build this table. The software can calculate Fraction, RSCU, RSCUmax, and Wi values automatically.
o How to access Kazusa Codon Usage web Database
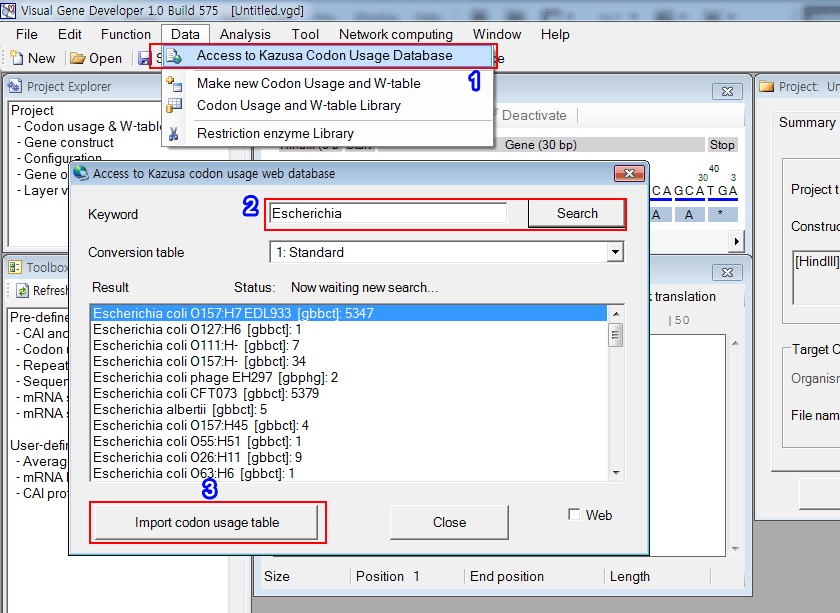
1. Click on the 'Access Kasusa Codon Usage Database' in the 'Data' menu
2. You can see the new window titled 'Access to Kazusa codon usage web database'. Enter a keyword and click on the 'Search' button
3. You can see the results in the list box. Click on the 'Import codon usage table' button
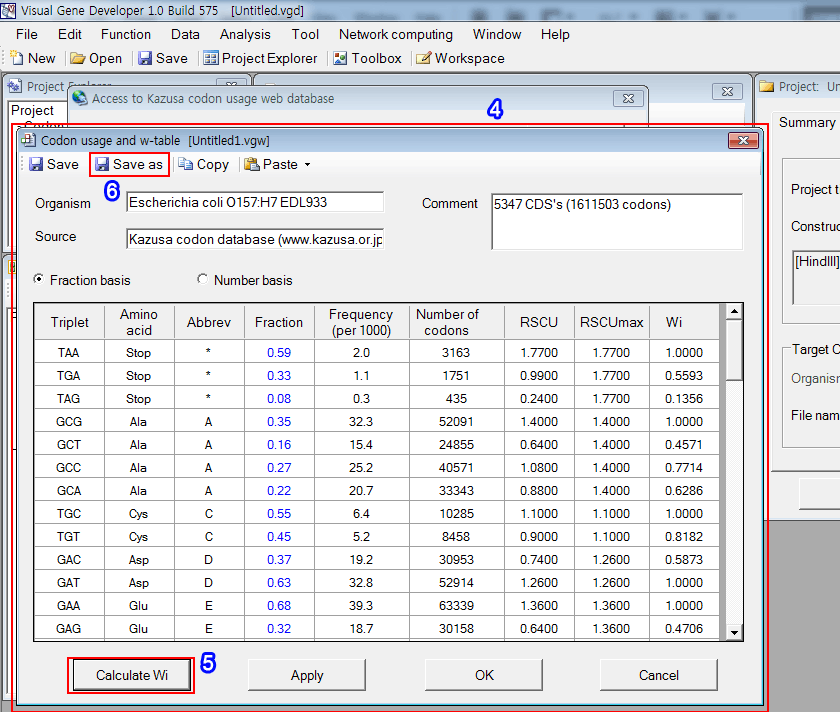
4. You can see the new window that has a codon usage table.
5. Further, click on the 'Calculate Wi' button to calculate other parameters as shown in the figure.
6. Save the current table clicking on the 'Save as' button.
o How to check all 'codon usage and w-table' files
1. Click on the 'Codon Usage and W-table Library' in the 'Data' menu
2. Sometimes, you may need to click on the 'Reload' button. But most of the case, it is not necessary. You can see the new update.
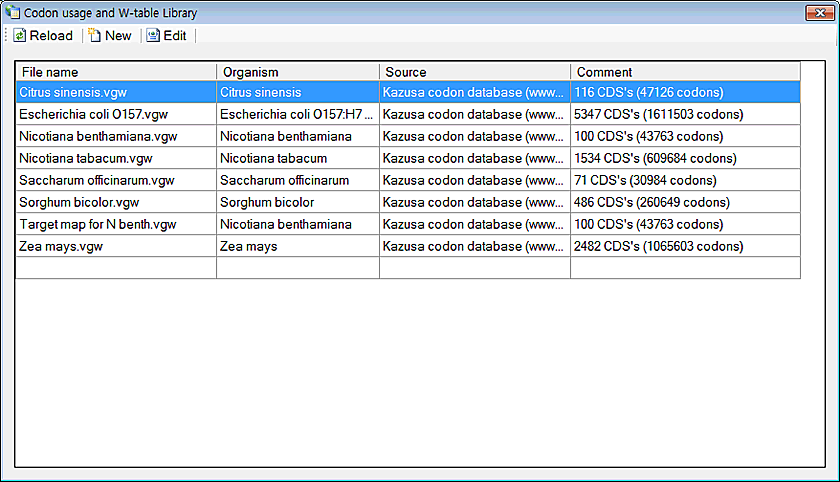
Information Visual Gene Developer automatically scans subfolder (/w-table) to find w-table files. Check the folder you installed Visual Gene Developer. Therefore, if you save files to another folder, our software will not show those files. Please save the w-table file to the default folder (/w-table).
o Access Genomic tRNA Database (http://gtrnadb.ucsc.edu/)