

Gene design
Visual Gene Developer optimizes a gene construct and generates a lot of candidate gene constructs. Therefore, the first step to optimize a gene is to define a gene construct. Usually, a gene construct consists of several different gene components. For example, a gene construct may have three components such as Start codon, Target gene, and Stop codon that is orderly connected like [Start codon]-[Target gene]-[Stop codon]. At the same, a gene construct can have only a single gene component like [Target gene]. In this case, a target gene may include both start codon and stop codon. Therefore, a gene construct contains at least one gene component.
In order to design a gene construct, the software has the 'Gene Construct Designer' window.
1. Click on the 'Gene construct' in the 'Project Explorer' window.
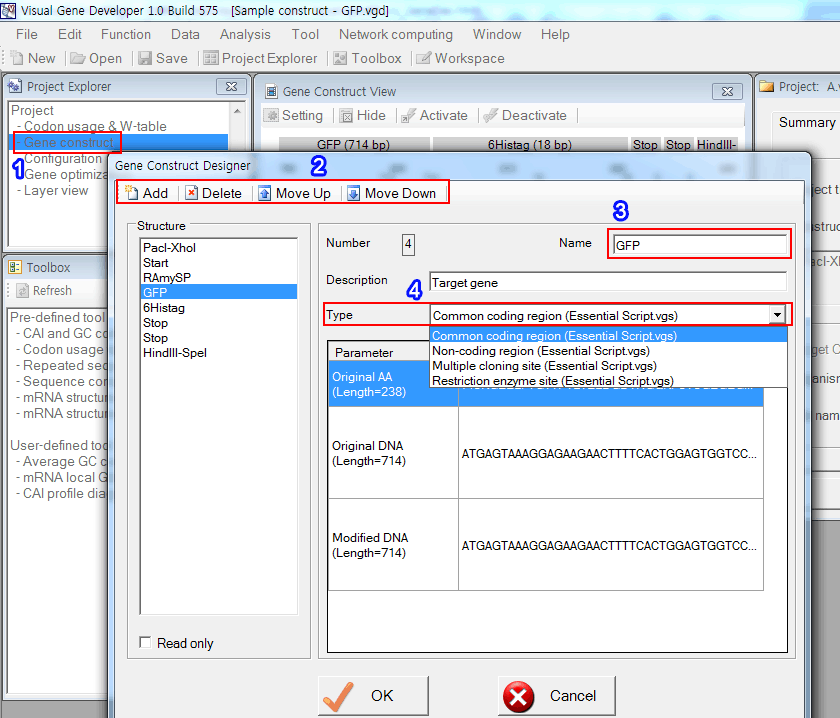
2. Click on the 'Add' button to add a new gene component, 'Delete' button to delete a current gene component, 'Move up' or 'Move down' button to change the order of gene components.
3. Since gene component names are frequently used in many other sections to represent target gene components, we recommend you using short names (below 10 letters).
4. There are four gene component types that are predefined in the 'Essential Script.vgs' file. Most users will use mainly the 'Common coding region' or 'Non-coding region'. Most implemented algorithms also make use of these two component types.
| Gene component type | Feature | Data format |
|
Common coding region |
Includes amino acid sequences |
Original AA: original amino acid sequence Original DNA: original DNA sequence Modified DNA: modified/optimized DNA sequence |
|
Non-coding region |
Pure DNA sequence |
Original DNA: original DNA sequence Modified DNA: modified/optimized DNA sequence |
|
Restriction enzyme site |
Single restriction enzyme site |
Original Format: string expression of restriction enzyme Original DNA: original DNA sequence Modified Format: modified/optimized string expression of restriction enzyme Modified DNA: modified/optimized DNA sequence |
|
Multiple cloning site |
Collection of restriction enzyme sites |
Original Format: string expression of restriction enzymes Original DNA: original DNA sequence Modified Format: modified/optimized string expression of restriction enzymes Modified DNA: modified/optimized DNA sequence |
As shown in the table, every gene component should have 'Modified DNA' as a data format parameter because Visual Gene Developer (specifically predefined algorithms) uses this parameter to optimize and analyze gene sequences. The optimization process will modify the DNA sequence of the 'Modified DNA' parameter value.
A user can define new gene component types like predefined ones. Check the 'Module development' section on the left menu.
