

Data conversion
Since Visual Gene Developer has easy copy & paste functions in many places, users can easily exchange data between the software and other commercial software such as NotePad, Excel, Word, SigmaPlot, Origin and etc. For example, a user may build an optimal codon usage table using Excel and then copy the table to Visual Gene Developer to make 'Codon usage and w-table'.
Moreover, our data files (Script, PropertyBag, codon usage, and w-table, gene construct file, neural network files) have a standard text format that can be easily editable using other editing software. Therefore, expert users or programmers can make their own modules to process (import/export) our Visual Gene Developer's data files (Check)
Regarding compatible data format, Visual Gene Developer can read DNA sequences from FASTA format file or extract field codes from GenBank format file (Check)
o Copy & Paste function
o Example 1: Gene construct or Codon Usage Calculation
1. Click on the 'Including full analysis result' menu to export the entire gene construct table to the clipboard. Paste it to Excel.
2. Click on the 'Copy' button. And then paste it to Excel.
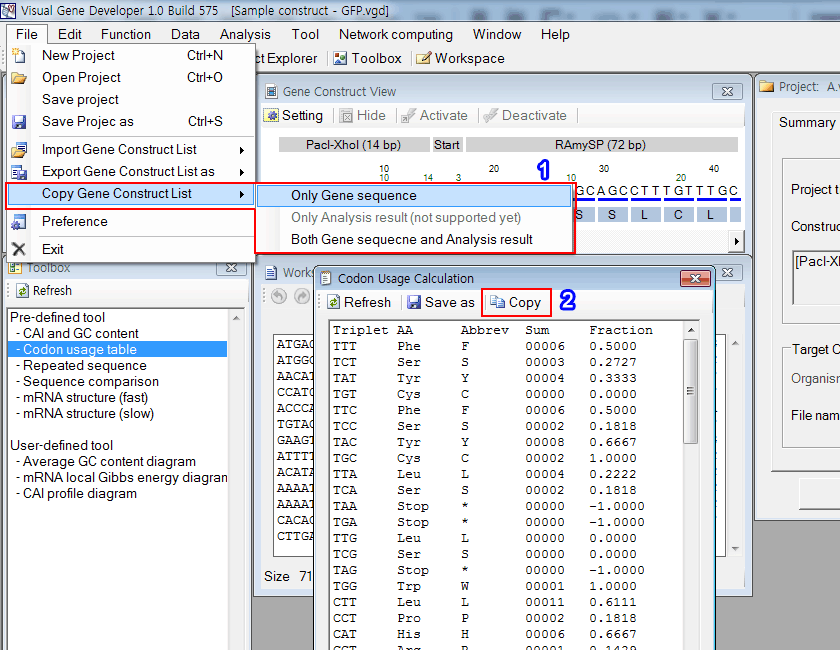
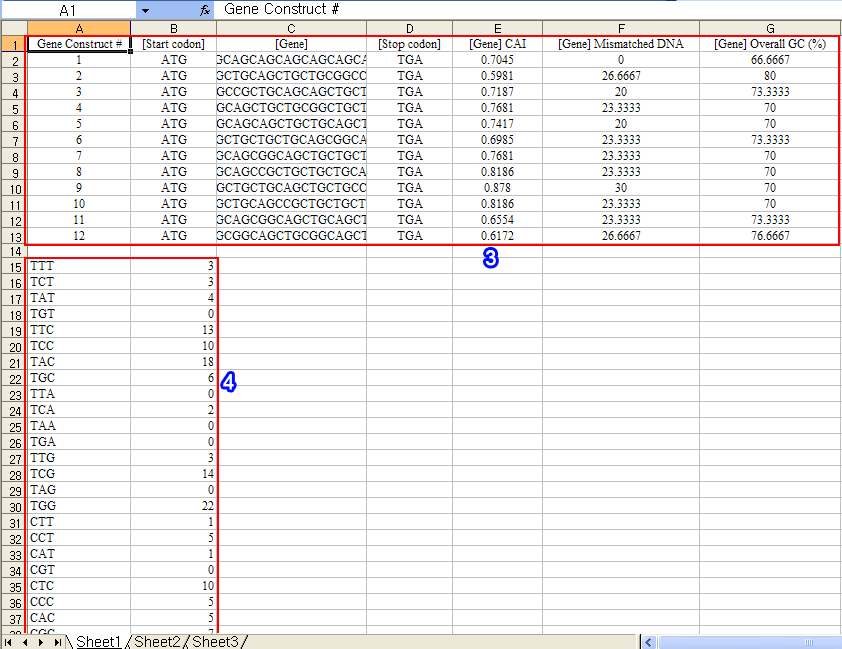
3. You can see the pasted gene construct table in the Excel document.
4. You can see the pasted codon usage table in the Excel document.
o Example 2: Paste codon usage table
1. Copy the region
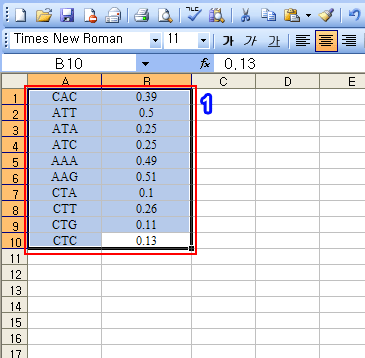
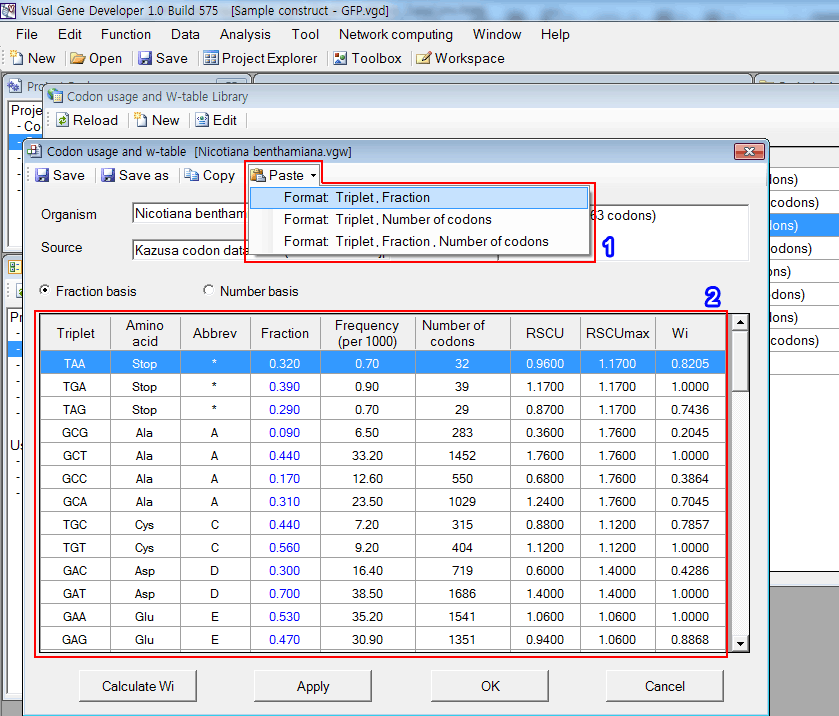
2. Click on the 'Format: Triple, Fraction' button
3. New data was pasted from the clipboard as shown in the red rectangular region.
o Import data from FASTA or GenBank file
- Import DNA sequences from FASTA file
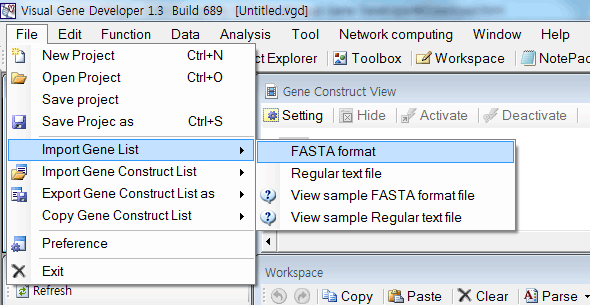
- Extract field information from GenBank file
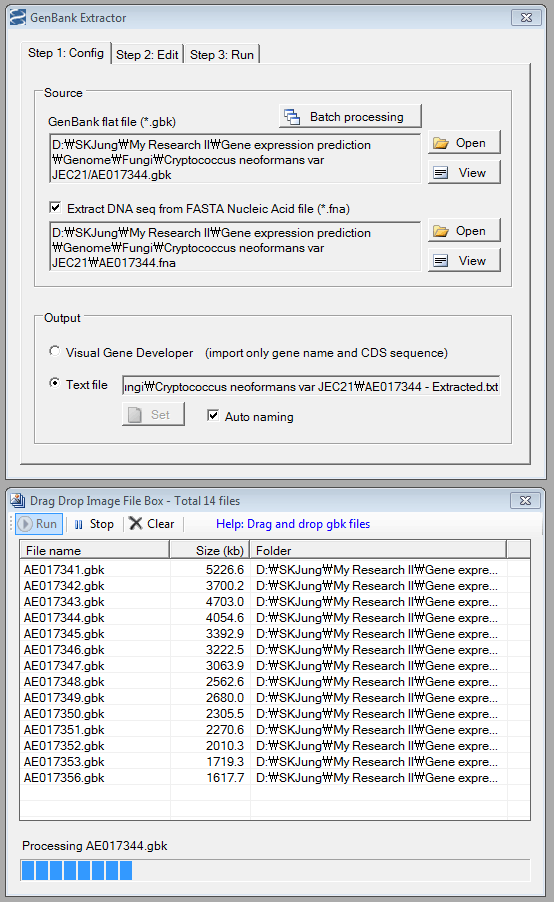
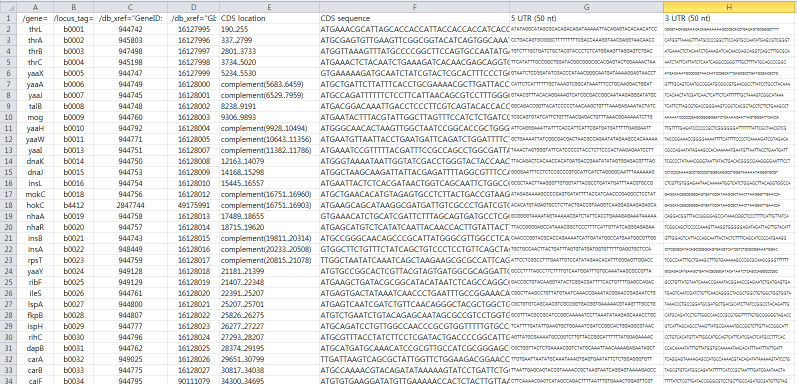
- Directly import Gene List from Clipboard
You can open data files using Windows NotePad.
o Project file (file extension: vgd)
Sample file. Project file has the most complicated data format. But easy to read.
|
<//Visual Gene Developer Project File//> |
o Script file (file extension: vgs)
Example file
|
<//Visual Gene Developer Script Language
File//> ACGCGAACAACGTGCCGGTACGGATCGCCGGCATCAACTGGTTTGGGTTCGAAACC
TGCAATTACGTCGTGCACGGTCTCTGGTCACGCGACTACCGCAGCA",True) |
Information: sequence should be single string.
o PropertyBag file (file extension: vgp)
Example file
|
<//Visual
Gene Developer PropertyBag
File//>
<//END OF COLLECTION//> |
o Codon usage and w-table file (file extension: vgw)
Example file
|
<//Visual Gene Developer W-table File//> |
o Neural network file (file extension: vgn)
Apparently, Neural network file format is slightly different from those of other data files. However, it is a higher standard INI text format.
|
[Application] |
