
Manipulation
This category is almost the same like 'Sequence optimization' or 'mRNA structure optimization'
We recommend using this category when a module just changes a sequence without optimization.
Information
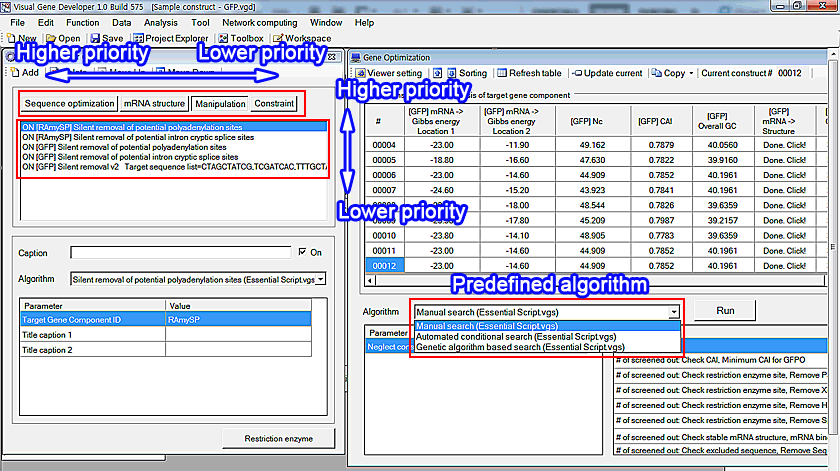
We provides two predefined 'Search strategy' algorithms (Manual search, Automated conditional search)
These two modules process 'Sequence optimization' first and then check 'mRNA structure optimization', 'Manipulation', and finally 'Constraint' modules. It means that there are different priorities.
o How to develop module
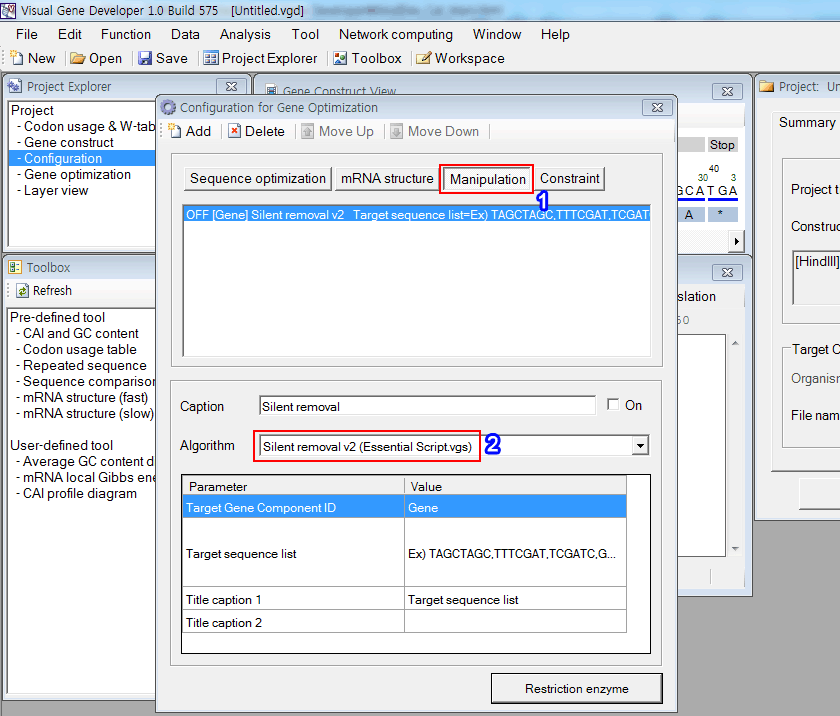
The following tutorial shows how we developed the 'Silent removal v2' module that is a predefined function.
1. When you click on the 'Manipulation' button in the 'Configuration of gene optimization' window
2. You will see the registered function: 'Silent removal (Essential Script.vgs)"
Step 1: Create module
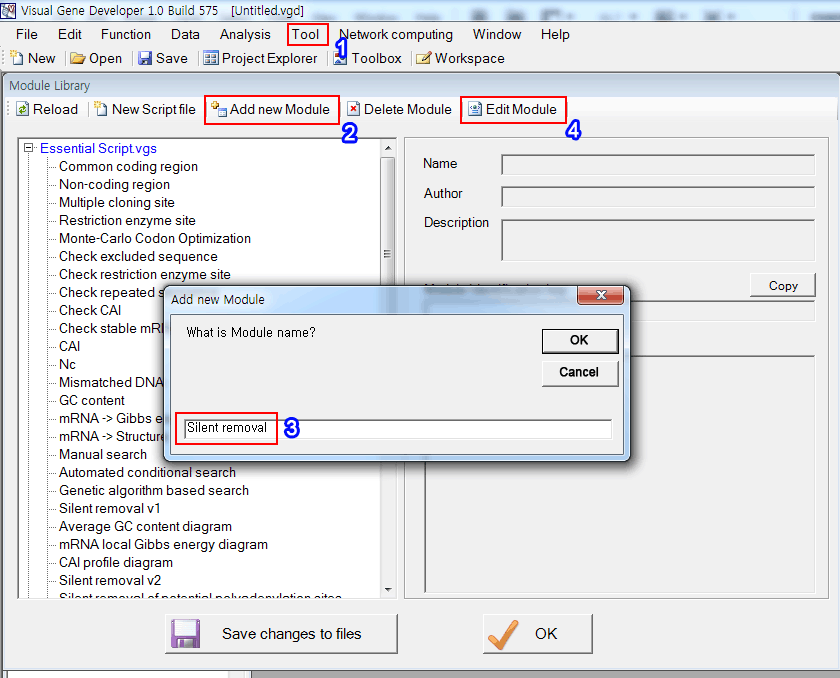
1. Click on the 'Module Library' in the 'Tool' menu
2. Click on the 'Add new Module' button to create a new module
3. Input module name
4. Click on the 'Edit Module' button
Step 2: Develop module
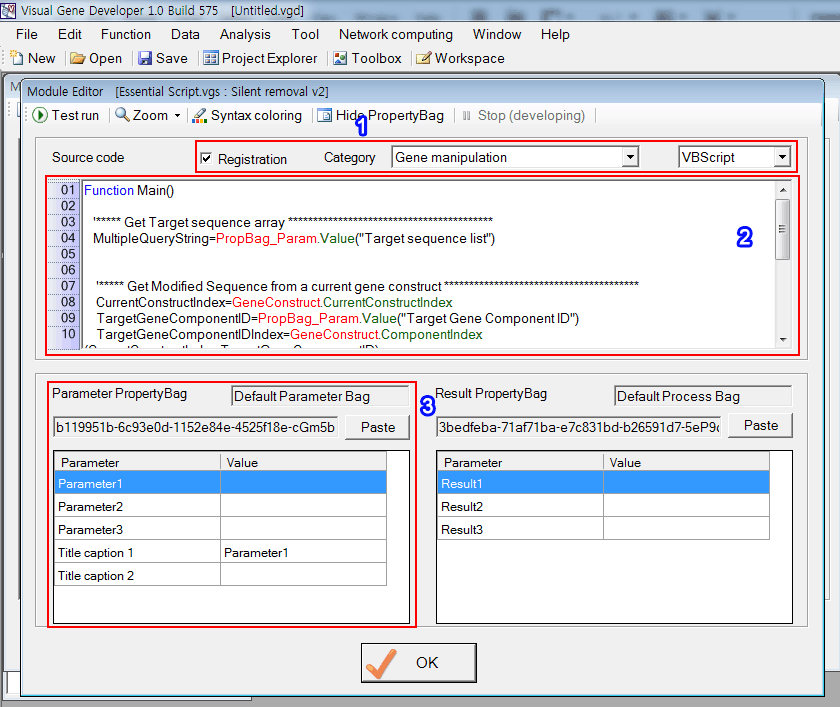
1. Set parameters as shown in the below figure.
2. Input source code
3. We will design custom PropertyBag to replace current PropertyBag
Here is the full source code (VBScript version). The Visual Gene Developer already included this module.
|
Function Main()
'***** Get Target sequence array ***************************************** MultipleQueryString=PropBag_Param.Value("Target sequence list")
'***** Get Modified Sequence from a current gene construct *************************************** CurrentConstructIndex=GeneConstruct.CurrentConstructIndex TargetGeneComponentID=PropBag_Param.Value("Target Gene Component ID") TargetGeneComponentIDIndex=GeneConstruct.ComponentIndex(CurrentConstructIndex,TargetGeneComponentID) OriginalSeq=GeneConstruct.ParameterValue(CurrentConstructIndex,TargetGeneComponentIDIndex, "Modified DNA")
'****** Perform Silent removal ********************************** ModifiedSeq=GeneService.Silent_Removal_MultipleQuery(OriginalSeq,MultipleQueryString) GeneConstruct.ParameterValue(CurrentConstructIndex,TargetGeneComponentIDIndex, "Modified DNA")=ModifiedSeq
End Function |
Step 3: Develop custom PropertyBag
1. Click on the 'PropertyBag Library' in the 'Tool' menu
2. Click on the 'Add new PropertyBag' button to create new one
3. Click on the 'Edit PropertyBag' button
4. To design the first parameter ('Target Gene component'), click on the the first line
5. Set parameters.
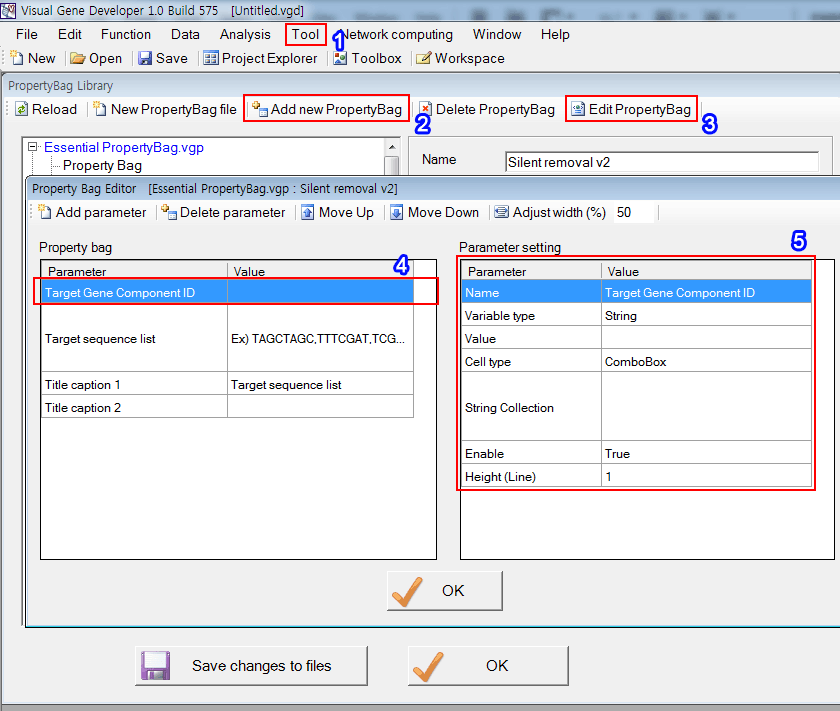
6. To design the second parameter ('Target sequence 1'), click on the the second line
7. Set parameters.
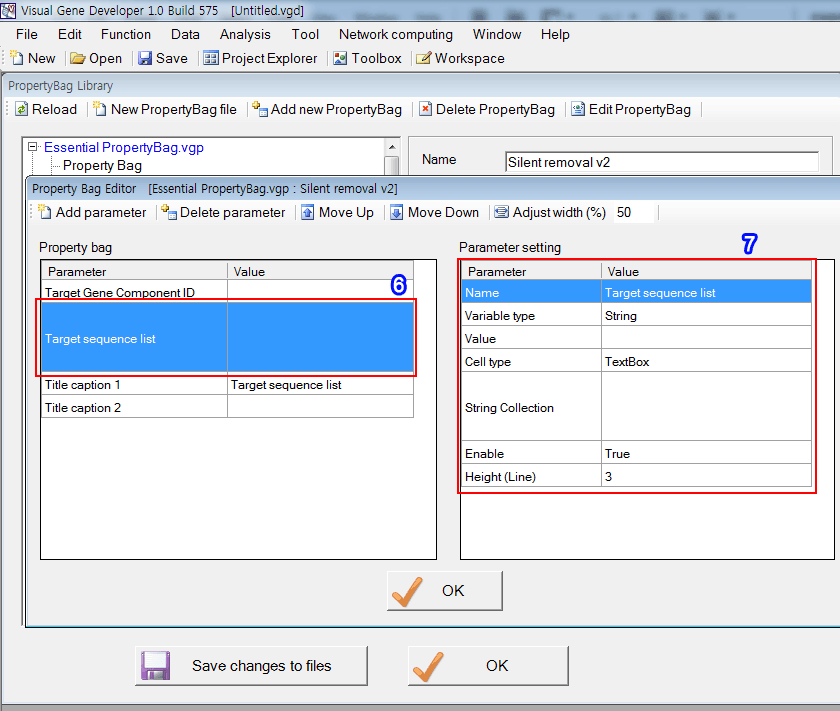
Step 4: Link PropertyBag to Module
8. Click on the 'Copy' button in the in the 'PropertyBag Library' window
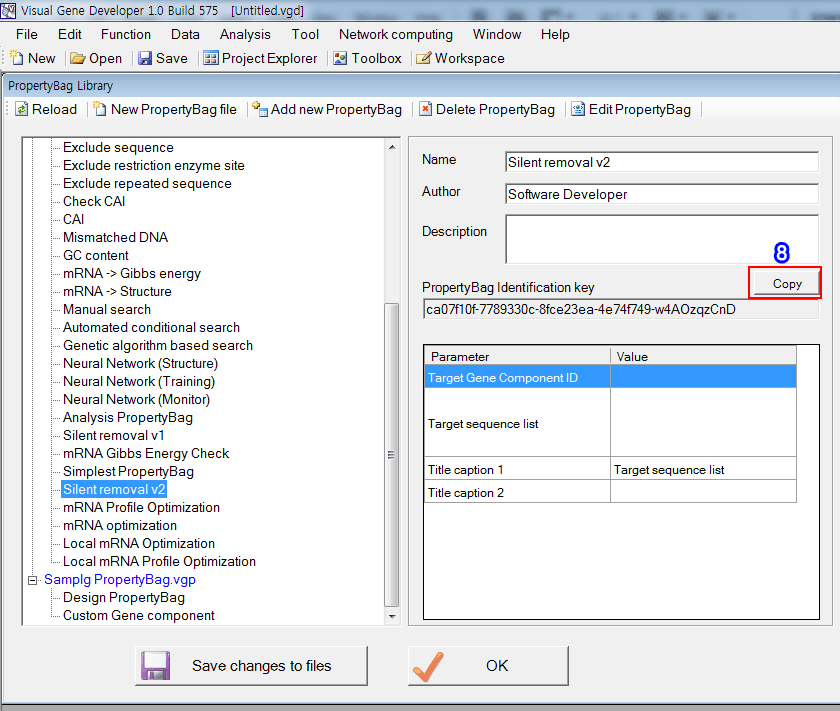
9. Click on the 'Paste' button in the 'Module Editor' window. You can see the imported PropertyBag
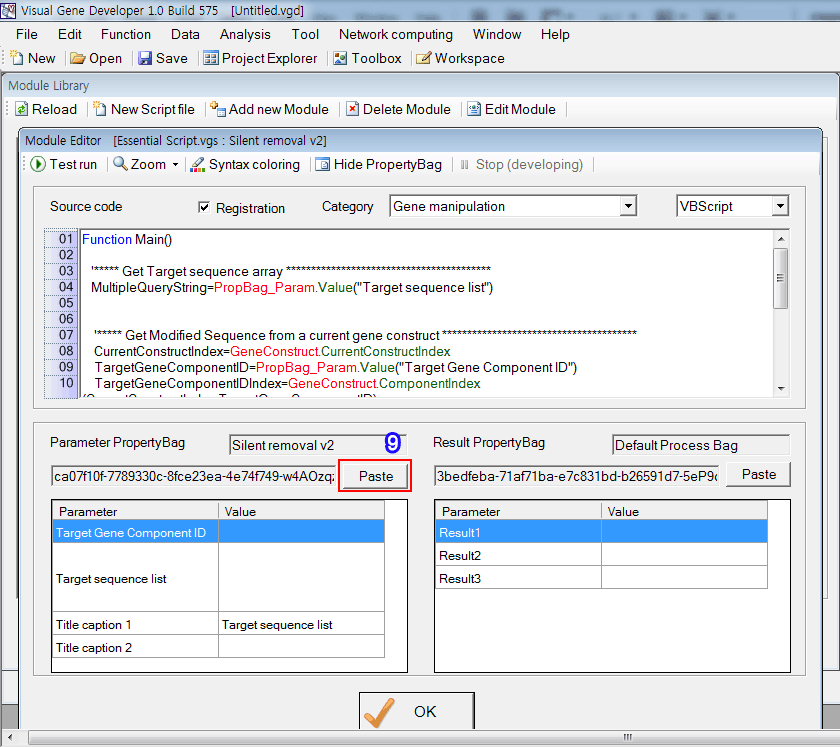
Step 5: Registration and application of module
1~2. You can see the module in the 'Configuration of gene optimization' window

