

Menu and user interface
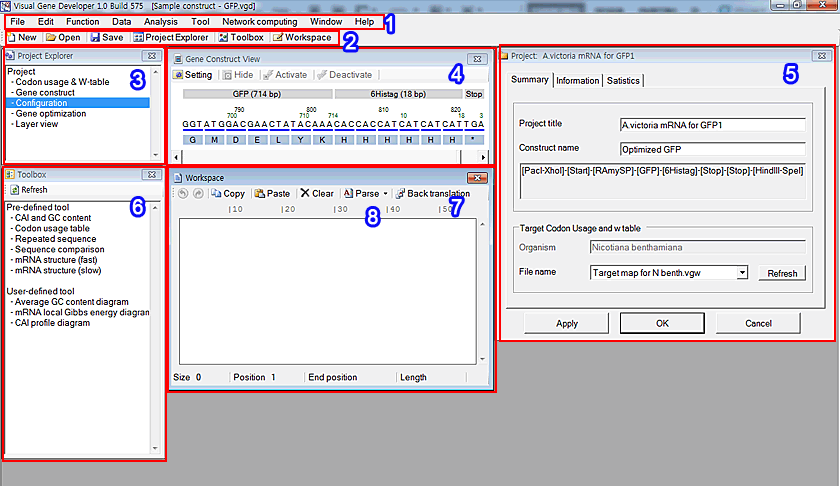
1. Main window menu: it includes most functions
2. Tool strip menu
3. Project Explorer: click on any item to show windows
4. Layer view manager: it shows information of a current gene construct graphically
5. Project window
6. Toolbox: click on any item to run it
7. Workspace: it contains a target sequence for gene analysis
8. Tool strip menu: almost all windows contain a localized tool strip menu
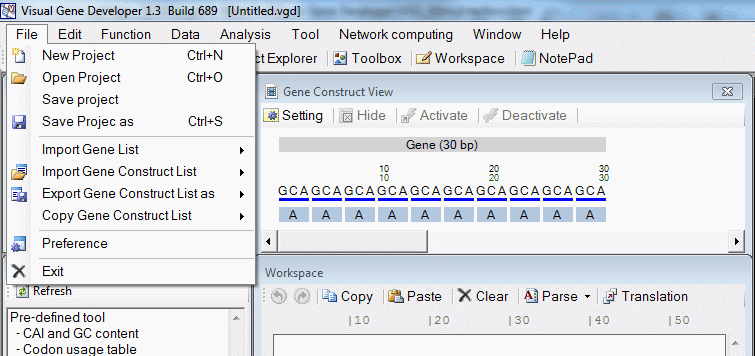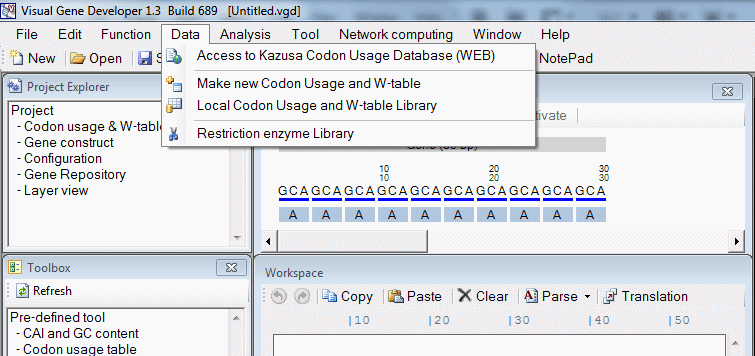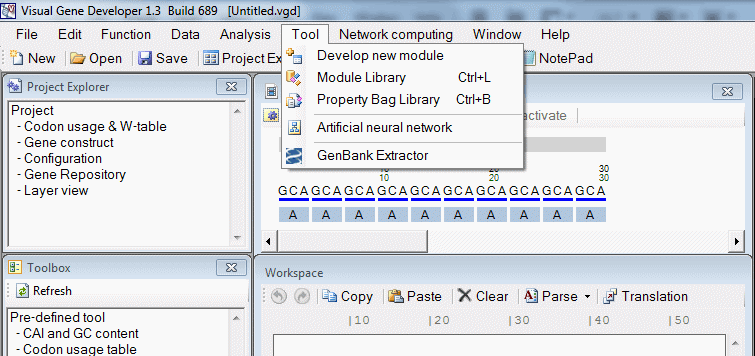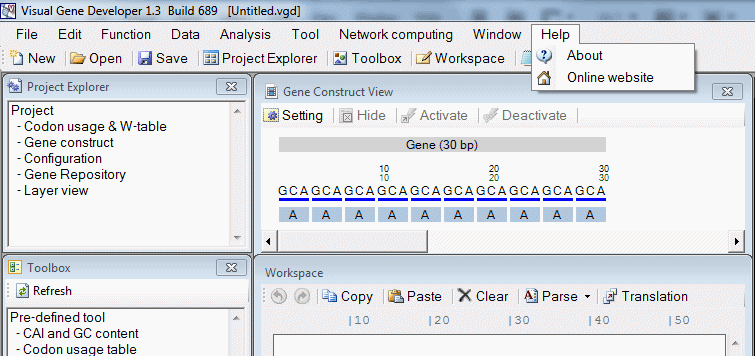
o Main window menu
Visual Gene Developer 1.3 (Build 689)
o Summary
| Main window menu | Description |
| File | Open and save data file |
| File > New project | Start new project |
| File > Open project | Open existing project |
| File > Save project | Save project file (file extension: vgp) |
| File > Save project as | Save project file as |
| File > Import Gene List > FASTA format | Import sequences from FASTA file |
| File > Import Gene List > Regular text file | Import sequences from regular text file |
| File > Import Gene List > View sample FASTA format file | |
| File > Import Gene List > View sample Regular text file | |
| File > Import Gene Construct List > Comma delimited standard text file | |
| File > Export gene construct as > Comma delimited standard text file | Export to standard text format file |
| File > Export gene construct as > Tab delimited standard text file | Export to standard text format file |
| File > Copy gene construct > Only gene sequence | Copy gene sequence to clipboard |
| File > Copy gene construct > Including full analysis result | Copy gene sequence to clipboard with analysis result |
| File > Preference | Configure environment |
| File > Exit | Exit |
| Edit | |
| Edit > Copy | Copy text of workspace window |
| Edit > Paste | Paste to workspace window |
| Edit > Search | Approximate sequence search (find similar sequences) |
| Edit > Delete all gene constructs except current one | Initialize construct list in the 'Gene optimization' window |
| Function | |
| Function > Parse sequence > Remove non-alphabetic characters | Parse text |
| Function > Parse sequence > To DNA sequence | Parse text |
| Function > Parse sequence > To Amino acid sequence | Parse text |
| Function > Formatting > Space delimited triplet | |
| Function > Formatting > Replace U with T | |
| Function > Formatting > Replace T with U | |
| Function > Formatting > Convert to upper case | |
| Function > Formatting > Convert to lower case | |
| Function > Complementary sequence | Make complimentary sequence |
| Function > Reverse sequence | Reverse source sequence |
| Function > Reverse complementary sequence | Make complimentary sequence |
| Function > Translate from DNA to amino acid | Translation |
| Function > Back translate from amino acid to DNA | Back translation |
| Function > Translate restriction enzyme | Convert restriction enzyme name into DNA sequence |
|
Function > Silent removal of sequences |
Perform silent removal of multiple sequences * Silent removal means a removal of certain target DNA sequence from a source sequence by replacing certain codons with synonymous codons while maintaining the expression of the same amino acids |
|
Function > Codon optimization |
Perform Monte-Carlo Codon Optimization (probability based random mutation) |
|
Function > mRNA optimization |
Perform mRNA Optimization combined with Codon Optimization |
|
Function > Degenerate gene sequence |
|
|
Function > File processing > Remove non-alphabetic letters in a file |
|
| Data | |
| Data > Access to Kazusa Codon Usage Database (WEB) | Access http://www.kazusa.or.jp/codon/ directly |
| Data > Access to Genomic tRNA Database (WEB) | Access http://gtrnadb.ucsc.edu/ directly |
| Data > Make new codon usage and w table | Create new w table file |
| Data > Lcal Codon usage and w table Library | Show w table library |
| Data > Restriction enzyme library | Show restriction library window |
| Analysis | |
| Analysis > Codon Usage Table | Build codon usage table |
|
Analysis > CAI, GC content, Nc |
Calculate several indexes such as CAI, GC content, Nc * CAI: Codon Adaptation Inex Nc: Number of effective codons |
|
Analysis > tRNA adaptation index (tAI) |
Calculate tAI |
| Analysis > Search sequence | Search multiple sequences |
| Analysis > Sequence comparison | Compare two sequences |
| Analysis > Repeated sequence | Search repeated sequence |
| Analysis > mRNA secondary structure | Calculate mRNA secondary structure |
| Analysis > Batch analysis mode | |
| Analysis > Gene optimization mode | |
| Tool | |
| Tool > Develop new module | Create new module |
| Tool > Module Library | Show module library |
| Tool > PropertyBag Library | Show PropertyBag library |
| Tool > Artificial neural network | Show artificial neural network window |
| Tool > GenBank Extractor | Extract field information from GenBank text file |
| Network computing | |
| Network computing > Server window | Show server control window |
| Network computing > Job list | Show current job status of network computing |
| Network computing > Add client | Add new clients |
| Network computing > Shutdown clients | Close all active clients |
| Window | |
| Window > Workspace | Show Workspace window |
| Window > Gene analysis | Show Gene analysis window |
| Window > Project window | Show Project window |
| Window > Gene construct designer | Show Gene construct designer window |
| Window > Configure gene optimization | Show Configuration of gene optimization window |
| Window > Gene optimization main window | Show Gene optimization window |
| Help | |
| Help > About | Show About window |
| Help > Online website | Direct link to our software website |
